
#PBP3 GENE RT QPCR FULL#
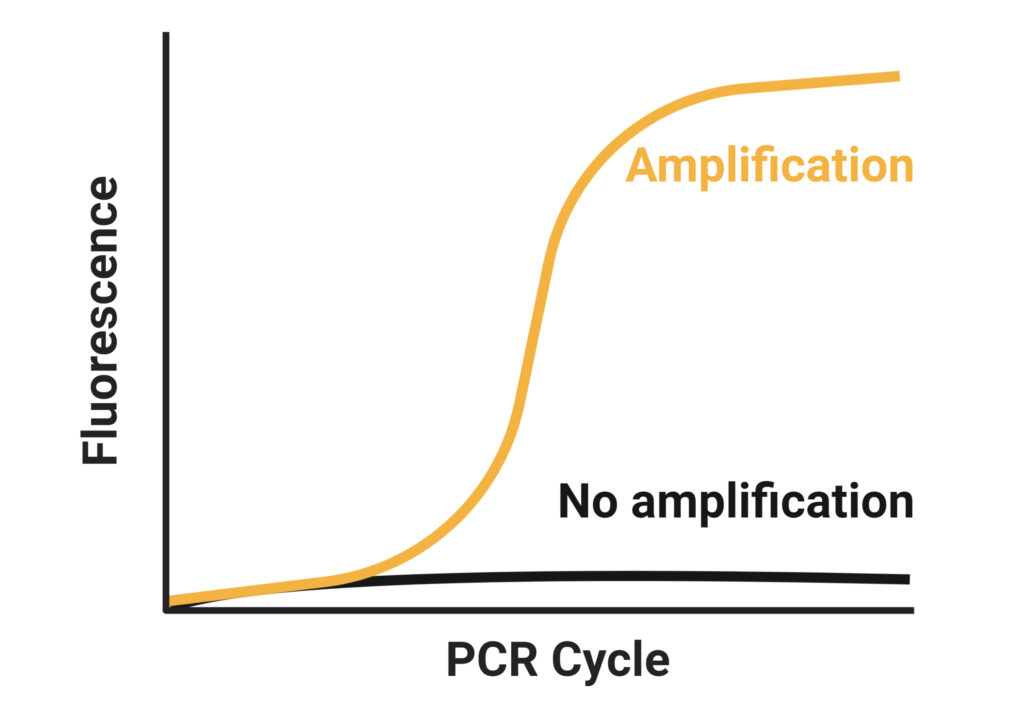
These primers anneal to the template mRNA strand and provide reverse transcriptase enzymes a starting point for synthesis. Often, a mixture of oligo(dT)s and random primers is used. Three different approaches can be used for priming cDNA reactions in two-step assays: oligo(dT) primers, random primers, or sequence specific primers ( Figure 2, Table 2). Taken together, total RNA is more suitable to use in most cases since relative quantification of the targets is more important for most applications than the absolute sensitivity of detection 1. Second, by avoiding any mRNA enrichment steps, one avoids the possibility of skewed results due to different recovery yields for different mRNAs.
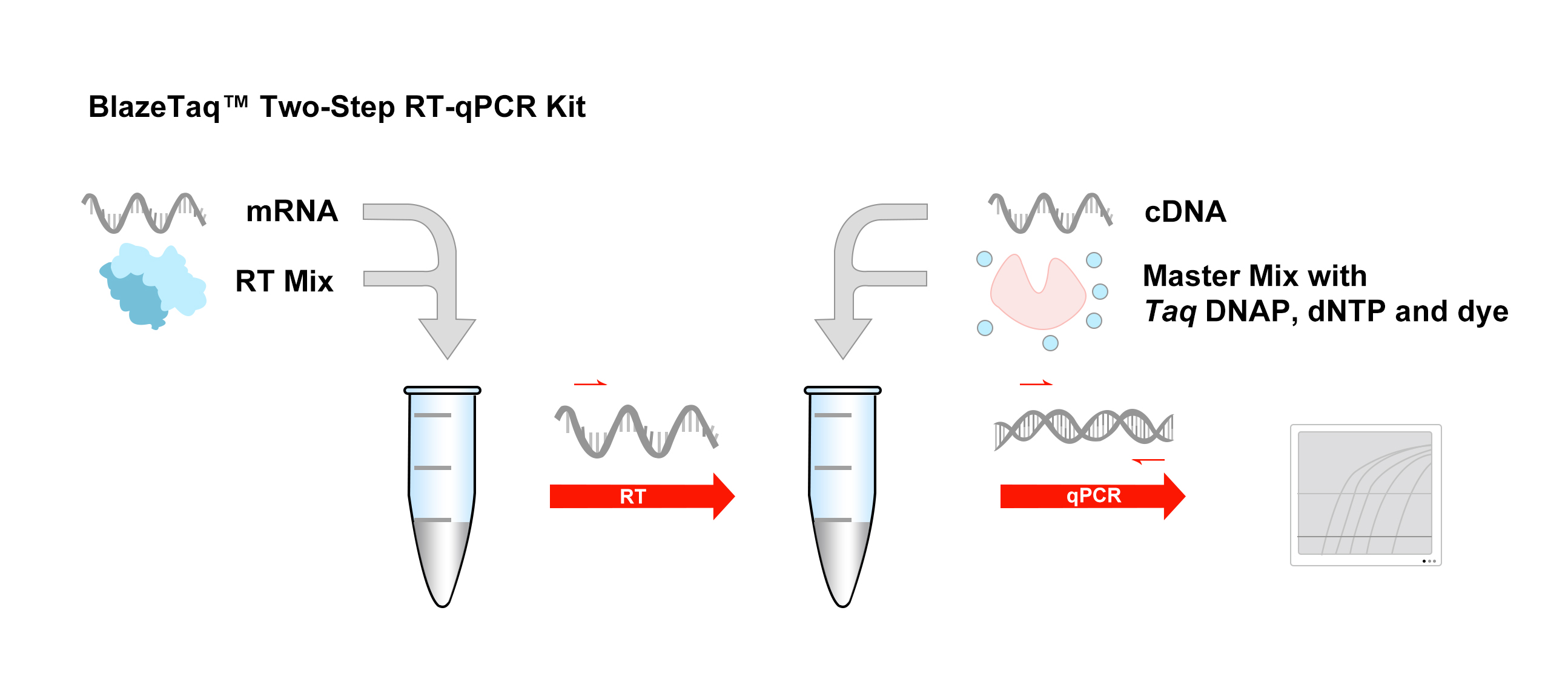
First, fewer purification steps are required, which ensures a more quantitative recovery of the template and a better ability to normalize the results to the starting number of cells. mRNA may provide slightly more sensitivity, but total RNA is often used because it has important advantages over mRNA as a starting material.

When designing a RT-qPCR assay it is important to decide whether to use total RNA or purified mRNA as the template for reverse transcription.


 0 kommentar(er)
0 kommentar(er)
